High Density SNP Mapping Used to Localize Type 1 Diabetes Susceptibility Alleles in Ancestral Haplotypes
Previous studies have revealed that the genetic predisposition for type 1 diabetes is strongly associated with the MHC chromosomal region, along with other regions located outside of the MHC class II which are also involved with the risk for developing the disease. However, it is generally recognized that the MHC class II loci are most likely to be involved directly with providing the risk factors causing diabetes.
Studies suggest that the MHC class II complex has the highest risk associated with DR3-DQ2 and DR4-DQ8 haplotypes. Other studies have also established that the DR3-DQ2 haplotypes do not exhibit an equal influence on the disease. AH8.1, also known as COX, and AH18.2, also known as QBL, are two of the conserved ancestral DR3-DQ2 haplotypes that have been known to be associated with type 1 diabetes. Both haplotypes contain the same HLA-DR and HLA-DQ alleles. However, the AH18.2 haplotypes seem to show a significant increase in risk when compared with other DR3-DQ2 containing haplotypes.
This study wanted to investigate and attempt to localize potential susceptibility alleles for type 1 diabetes present within the ancestral AH18.2 haplotype, through the use of high density SNP mapping. They accomplished this by analyzing the AH18.2 haplotypes of 10 selected DR3/TNFa1b5 homozygous subjects chosen from the Spanish population (8 were type 1 diabetics and 2 were controls), in order to determine the conservation of the AH18.2 haplotype. All samples collected were genotyped for HLA-DRB1 alleles as well as 2 microsatellie markers, TNFa and TNFb, which were established through the use of ABI Prism 3100 automatic sequencer. Comparisons were then made between these conserved regions and other DR3 haplotypes (i.e. AH8.1) and through the use of the Illumina’s HumanHap550 Bead chip they were able to execute a wide-range scan of genotypes for the entire MHC region.
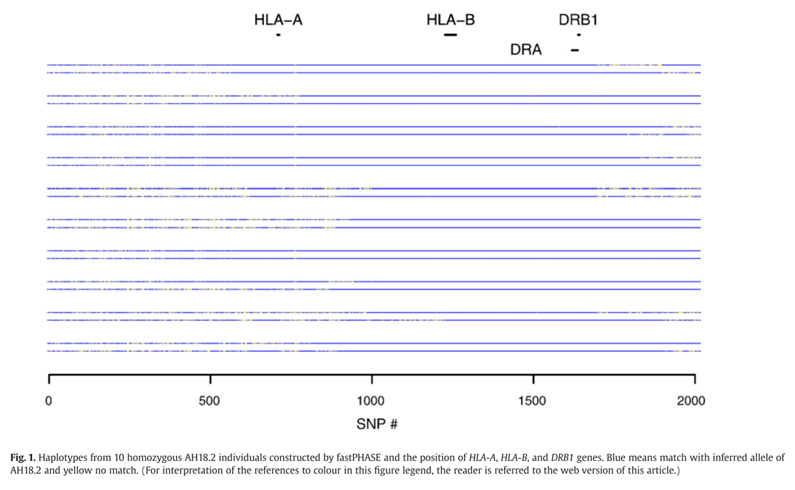
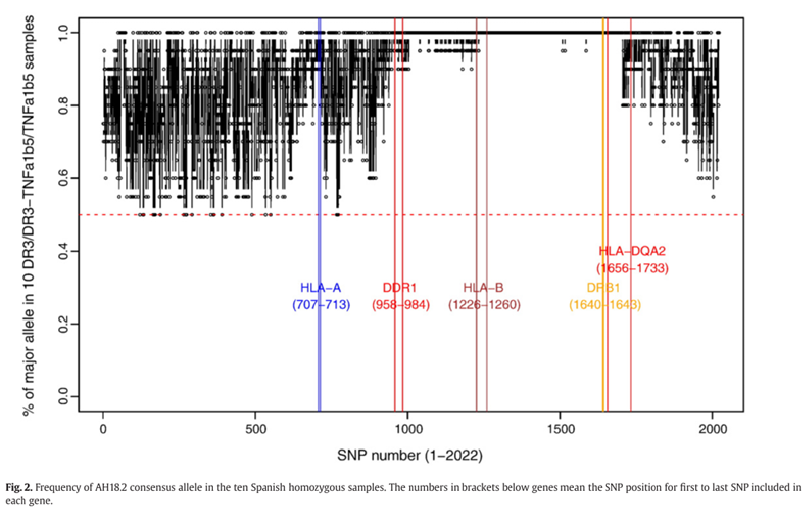
They discovered that matches, shown in blue on figure 1, were highest between regions centromeric to HLA-A genes and telomeric to the HLA-DRB1 genes. Moreover, they discovered that the frequency of the heterozygous samples collected of the AH18.2 alleles were near 1 for SNPs within both the DDR1 and HLA-DQA1 gene regions, as seen in figure 2. Meaning that AH18.2 is highly conserved between those two genes and a possible susceptibility gene may be located within this region.
They then conducted an analysis of 20 individuals from the U.S. Caucasian population that were homozygous for the AH8.1 allele. They observed similar results to the AH18.2 patterns, which showed a significant number of subjects, were homozygous across the whole MHC region.
With previous data suggesting that the AH18.2 haplotype possesses further risk alleles which are not present within the AH8.1, they then characterized and compared the SNP alleles present in these haplotypes for all genes located between DDR1 and HLA-DRA. As they grouped the 646 SNPs that have been studied within this region by gene, they found 68 genes in total which mapped between DDR1 and HLA-DRA genes. 18 of the genes were located in class I, 48 in class III, and only 2 were located in class II. All of these genes were found to be identical amongst the two haplotypes, and all the alleles found in HLA-DRB1 and HLA-DQA1 were the same in AH18.2 and AH8.1. However, through their full comparison of both the AH18.2 and the AH8.1 haplotypes, they found that within this conserved region between DDR1 and HLA-DRA1, 233 SNPs were different. Suggesting that inside this MHC region present between the DDR1 and HLA-DRA genes there are other susceptibility alleles for type 1 diabetes within the AH18.2 haplotype.
| Gene | % of different SNPs |
| DDR1 | 28.00 |
| VARSL | 71.43 |
| SFTPG | 40.00 |
| DPCR1 | 50.00 |
| MUC21 | 28.57 |
| C6orf15 | 26.92 |
| CDSN | 28.57 |
| PSORS1C1 | 61.54 |
| PSORS1C2 | 33.33 |
| CCHCR1 | 56.25 |
| TCF19 | 75.00 |
| POU5F1 | 40.00 |
| HCG27 | 36.84 |
| HLA-C | 42.86 |
| HLA-B | 34.38 |
| MICA | 40.00 |
| HCP5 | 22.86 |
| MICB | 41.18 |
| MCCD1 | 42.86 |
| BAT1 | 66.67 |
| ATP6V1G2 | 0.00 |
| NFKBIL1 | 28.57 |
| LTA | 33.33 |
| TNF | 100.00 |
| LST1 | 0.00 |
| NCR3 | 20.00 |
| AIF1 | 0.00 |
| BAT2 | 0.00 |
| BAT3 | 12.50 |
| C6orf47 | 50.00 |
| BAT4 | 0.00 |
| LY6G5B | 33.33 |
| LY6G5C | 0.00 |
| BAT5 | 50.00 |
| LY6G6F | 50.00 |
| LY6G6E | 100.00 |
| LY6G6D | 0.00 |
| LY6G6C | 50.00 |
| MSH5 | 25.00 |
| C6orf27 | 0.00 |
| LSM2 | 100.00 |
| HSPA1L | 0.00 |
| HSPA1B | 0.00 |
| C6orf48 | 33.33 |
| NEU1 | 0.00 |
| SLC44A4 | 0.00 |
| EHMT2 | 33.33 |
| ZBTB12 | 100.00 |
| C2 | 0.00 |
| CFB | 33.33 |
| RDBP | 100.00 |
| SKIV2L | 42.86 |
| STK19 | 33.33 |
| TNXB | 7.69 |
| ATF6B | 0.00 |
| PRRT1 | 0.00 |
| PPT2 | 0.00 |
| EGFL8 | 100.00 |
| AGPAT1 | 50.00 |
| RNF5 | 50.00 |
| AGER | 33.33 |
| PBX2 | 100.00 |
| GPSM3 | 50.00 |
| NOTCH4 | 36.36 |
| LOC401252 | 50.00 |
| C6orf10 | 34.15 |
| BTNL2 | 37.04 |
| HLA-DRA | 50.00 |
Table 1. Percentage of different SNPs between AH18.2 and AH8.1 in the 68 genes that map in the MHC region between DDR1 and HLA-DRA genes.
This study concluded that the MHC is one of the most important susceptibility regions when it comes to autoimmune diseases, as well as genetically contributing largely to type 1 diabetes risk. The AH18.2 haplotype has also been known as the “diabetogenic haplotype” due to its significant involvement with increasing risk for type 1 diabetes, in comparison to the other haplotypes that possess the same HLA-DRB1 and DQ alleles. The scientists who conducted this study stated, “The high susceptibility must be due to a gene or genes different from the HLA class II loci and one of these genes appears to be the HLA-B18 allele. However, we wish to raise the possibility that there may be additional susceptibility alleles on this haplotype… Our study allows us to exclude the regions telomeric to DDR1 and centromeric to HLA-DRA as harboring the additional susceptibility factors.”
The overall sequence difference of these two haplotypes have been reported to involve 15,345 SNPs within the MHC region, and around 40% of these SNPs exhibited differences between the two haplotypes. This studies data was nowhere near as extensive as evaluating all 15,345 involved SNPs within the MHC region. However, it did lay a good foundation, to initiate further studies on the significant number of genes and polymorphisms left to explore within the AH18.2 haplotype, and/or further studies for expanding on the suspected MHC region likely to possess these additional susceptibility alleles.
References
Santiago JL, Li W, Lee A, Martinez A, Chandrasekaran A, Fernandez-Arquero M, Khalili H, Concha EGDL, Urcelay E, Gregersen PK. Localization of Type 1 Diabetes susceptibility in the ancestral haplotype 182 by high density SNP mapping. Genomics. 2009 Jul 8;94(4): 228-232.
Figure images linked to Journal URL
- Applications of the ‘Omics fields in Type 1-Insulin Dependent Diabetes Mellitus (IDDM1) Research
- Understanding pancreatic beta-cell death in type 1 diabetes through the use of systems biology