Applications of the ‘Omics fields in Type 1-Insulin Dependent Diabetes Mellitus (IDDM1) Research
I will be covering how Genomics, Proteomics and other ‘Omics fields are involved in the research and discovery of new knowledge essential for our further understanding of Type 1- Insulin Dependent Diabetes Mellitus (IDDM1). These blogs will explore the discoveries and how they have led and continue to lead us towards better understanding this complex disease. I believe that through these different fields of research we, as scientists, could fundamentally uncover crucial information necessary for finding better treatments for those affected and ultimately revealing its cure. However, in order to accomplish this, we still have a great deal of research ahead. IDDM1 is a complex disease, where many factors play a role, most of which have yet to be determined.
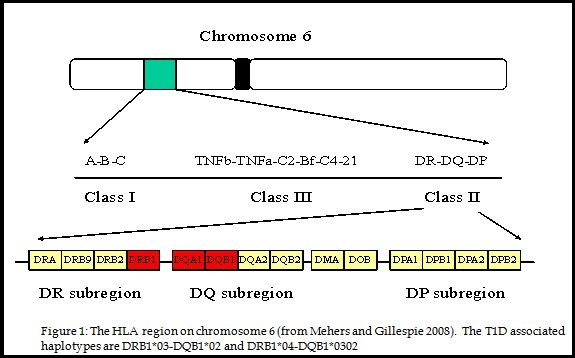
A little overview of Type 1- Insulin Dependent Diabetes Mellitus (IDDM1), it is a complex, autoimmune disease, which causes the destruction of insulin producing beta cells in the pancreatic islets. (McCulloch et al. 2014) The inheritance pattern of IDDM1 is still unknown. It is understood that a predisposition for developing IDDM1 is passed through generations within families, but identifying this pattern is complicated due to the many different genes and factors in which influence a person’s risk for developing this disorder. These factors involved include peptides, proteins, metabolites, nucleic acids, and polymorphisms, each of these have been recognized as biomarkers for IDDM1. So far, there have been around 18 to 20 regions within the genome that have been linked to influencing the risk for IDDM1. (Adams et al. 2011) One of these regions found to have an involvement is the HLA complex. Variations of the HLA genes have been found to play an important role in being genetic risk factors, but alone are not causal for the disease, other genetic factors, along with non-genetic factors are also involved. Some environmental factors are being research as triggers for those who have these risk factors, these include, colder weather/climates, viruses, along with early diet regiments. (Knip and Simell et al. 2012)
Since so much is yet to be discovered on the causal elements of IDDM1, many ‘Omics fields are working on producing effective research to find these answers. Here are some examples of these specific fields and how they are applied towards IDDM1 research, along with links to some of this research,
Proteomic techniques are extremely valuable for improving our knowledge on beta-cell function in IDDM1, as well as the search towards therapeutic targets.
http://dvr.sagepub.com/content/2/2/54.full.pdf+html
http://www.mcponline.org/content/4/4/441.full
Transcriptomics is the study of the complete set of RNA transcripts that are produced by the genome, under specific circumstances or in specific cells. This field could gather more information on the gene expression of the human pancreas as well as purified islets in IDDM1 patients
Immunomics is the study of immune system regulation and its response to pathogens. Further research from this field would provide more knowledge on the antibodies involved in the destruction of beta cells.
http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.0020081
Metabolomics is an approach used to identify and assess metabolic characteristics, changes, and phenotypes in response to influencing factors, such as environment, diet, and lifestyle. With IDDM1 having multiple involved factors, including the environment, this field may be able to uncover which factors are fundamental and how they affect the metabolic pathways of IDDM1 patients.
http://www.medscape.com/viewarticle/754507
Pharmacogenomics is the study of how genes affect ones response to drug treatments, which is a huge contributor towards personalized medicine. This field could lead to further discoveries on insulin and possibly better therapies, such as the use of regulatory T-cells, and more.
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2646064/
Stem Cell Genomics is a study which analyzes the genome of stem cells. Applications of research within this field could lead to new drug discoveries and provide information on diseases by the molecular characterizations through DNA and transcriptome sequencing and through exploring the epigenetic changes of stem cells and their subsequent products.
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3239236/
System Biology can assist us in understanding the regulation of involved gene networks, and how they control beta cell death, as well as the involvement with the ‘dialogue’ between beta cells and the immune system.
http://www.numesi.com/inscription/EIZIRIK-1.pdf
These fields are also laying the grounds for discovering new innovative bioengineered devises, giving IDDM1 patients more control over the disease and provide them with tools that could help them live a more normal life. An example of this is the new research on nanosensors and nanomaterials used for monitoring glucose levels in diabetics.
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2996880/
(Image 2) http://www.discoverymedicine.com/Didier-Hober/2010/08/27/enteroviral-pathogenesis-of-type-1-diabetes/
(Image 1) http://www.intechopen.com/source/html/24180/media/image1.jpg
References
Adams A. Genes Can Cause Type 1 Diabetes Walston J, Silver K. Genes Can Cause Type 1 Diabetes. 2011.
Braga-Neto UM, Marques ETA. From Functional Genomics to Functional Immunomics: New Challenges, Old Problems, Big Rewards. PLoS Computational Biology. 2006;2(7): e81.
Cash KJ, Clark HA. Nanosensors and nanomaterials for monitoring glucose in diabetes. Trends in Molecular Medicine. 2010;16(12): 584-593.
Eizirik D, Moore F, Flamez D, Ortis F. Use of a systems biology approach to understand pancreatic β-cell death in Type 1 diabetes. Biochemical Society Transactions. 2008 1;36(3): 321.
Jin Y, She J. Novel Biomarkers in Type 1 Diabetes. The Review of Diabetic Studies. 2012;9(4): 224-235.
Knip M, Simell O. Environmental Triggers of Type 1 Diabetes. Cold Spring Harbor Perspectives in Medicine. 2012 May 23;2(7): a007690-a007690.
McCulloch DK. Patient information: Diabetes mellitus type 1: Overview (Beyond the Basics) Nathan DM, Mulder JE. Diabetes mellitus type 1: Overview. 2014 Sep 15.
Mckillop AM, Flatt PR. Emerging Applications of Metabolomic and Genomic Profiling in Diabetic Clinical Medicine. Diabetes Care. 2011 21;34(12): 2624-2630.
Putnam AL, Brusko TM, Lee MR, Liu W, Szot GL, Ghosh T, Atkinson MA, Bluestone JA. Expansion of Human Regulatory T-Cells From Patients With Type 1 Diabetes. Diabetes. 2008 3;58(3): 652-662.
Scott EM, Carter AM, Findlay JBC. The application of proteomics to diabetes. Diabetes & vascular disease research : official journal of the International Society of Diabetes and Vascular Disease. 2005;2(2): 54.
Sparre T. Unraveling the Pathogenesis of Type 1 Diabetes with Proteomics: Present And Future Directions. Molecular & Cellular Proteomics. 2005 25;4(4): 441-457.
Weir GC, Cavelti-Weder C, Bonner-Weir S. Stem cell approaches for diabetes: towards beta cell replacement. Genome Medicine. 2011;3(9): 61.


One thought on “Applications of the ‘Omics fields in Type 1-Insulin Dependent Diabetes Mellitus (IDDM1) Research”